
The canonical function of aminoacyl-tRNA synthetase (aaRSs) is to catalyze the aminoacylation reaction of their cognate tRNAs to form aa-tRNAs, which are fundamental materials for protein biosynthesis. Compared with existed as free form in prokaryotes or low eukaryotes, aaRSs in higher eukaryotes could form various aminoacyl-tRNA synthetase complexs through interactions mediated by their progressive domains obained in evolution, such as the MSC formed by nine aaRSs and three scaffold proteins in cytoplasm of mammalian. Recent results studying on aaRSs have shown that MSC components could be released out of MSC quickly induced by enzyme cascade reactions under outer signal stimulation, and further participate in other biological processes,different from their canonical function (The release of members from MSC mainly relies on the phosphorylation modification on their key amino acids by protein kinases). Overall regulation on the MSC is rarely reported. Therefore, finding the key factors involving in overall regulation on the MSC would reveal the molecular mechanism of regulating protein biosynthesis and also provide fundamental theory for the investigation into non-canonical functions of aaRSs or the diagnosis and cure for aaRSs-related diseases.
Under the guidance of Prof. WANG Enduo at the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, graduate students LEI Huiyan et al identified calpain 2 which is Calcium-Activated Neutral Proteinase 2, interacting with leucyl-tRNA synthetase (LeuRS) through yeast two-hybrid screen.
Further studies showed that both calpain 1 and calpain 2 could partially hydrolyze most MSC components, affect their incorporations into the MSC and generate specific fragments that resembled those previously reported. The cleavage sites of calpain in ArgRS, GlnRS and p43 were precisely identified. After cleavage, their N-terminal regions were removed. Sixty-three amino acid residues were removed from the N-terminus of ArgRS to form ArgRSΔN63. GlnRS formed GlnRSΔN198 and p43 formed p43ΔN106. The enzymatic activities of the truncated ArgRS showed little effect as compared with those of the wild-type enzyme; however, GlnRSΔN198 had a much weaker affinity with its substrates, tRNAGln and glutamine, indicating that calpain cleavage might have great impact on the glutamine-dependent anti-apoptotic activity of GlnRS reported by other lab.
Furthermore, they showed that p43ΔN106, which was exactly the same with p43(ARF) (Apoptosis-Released Factor) reported previously, could be abundantly produced in U937 cells and further secreted into culture medium under FBS-starvation conditions. The formation and secretion of p43ΔN106 in U937 mainly depended on the hydrolytic activity of calpain.
These results showed, for the first time, that calpain plays an essential role in dissociating the MSC and might regulate the canonical and non-canonical functions of certain components of the MSC.
This work, entitled "Calpain cleaves most components in the multiple aminoacyl-tRNA synthetase complex and affects their functions", was published online in Journal of Biological Chemistry on August 31th, 2015. This work is funded by the National Natural Science Foundation of China, the Ministry of Science and Technology of China, the Chinese Academy of Sciences and the Science and Technology Commission of Shanghai Municipality.
CONTACT:
WANG Enduo, Principal Investigator
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai 200031, P. R. China
Phone: 86-21-54921241
E-mail: edwang@sibcb.ac.cn
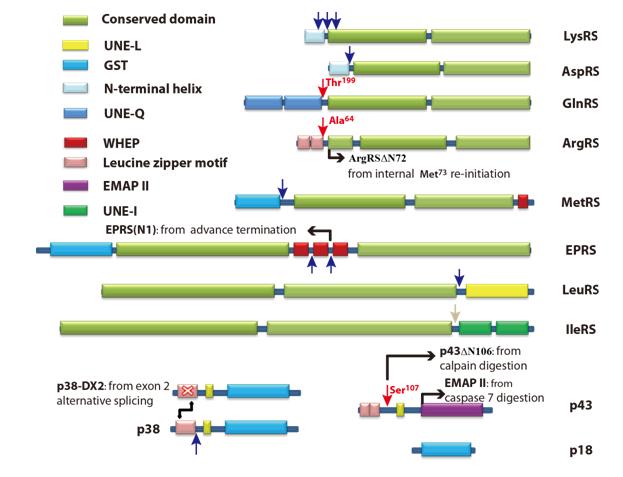
Sketch map: Proposed locations of calpain cleavage sites of MSC components (Image by Prof. WANG Enduo`s group)

86-10-68597521 (day)
86-10-68597289 (night)

86-10-68511095 (day)
86-10-68512458 (night)

cas_en@cas.cn

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

