
In a study published in Cell, a research team led by ZHANG Yong'e and WANG Haoyi from the Institute of Zoology of the Chinese Academy of Sciences has characterized the diversity of DNA transposons and expanded the genome engineering toolbox.
"Our systematic and comparative framework complements conventional case studies in elucidating basic biology and strengthening applied biology," said Prof. ZHANG, corresponding author of the study.
Since the first discovery of transposons by Barbara McClintock in the 1940s, scientists have been fascinated by these "jumping genes" and their role in evolution. As one of the major transposon types, DNA transposons have received considerable attention. However, due to the limited scale of previous case studies, the factors associated with transposition activity and evolutionary patterns have remained unclear.
In this study, the researchers predicted 130 potentially active DNA transposons from 102 animal genomes. Through experimental screening and validation, they identified 40 novel transposons with activity in human cells, increasing the number of active transposon vectors in mammals from 20 to 60 and significantly expanding their evolutionary diversity.
Taking advantage of the largest dataset of active DNA transposons ever obtained through systematic experimental screening, they performed further in-depth analyses, deciphering factors underlying transposition activity, exploring evolutionary dynamics, and identifying diverse functional properties.
One notable finding is that the Tc1/mariner superfamily exhibits enhanced activity, which underlies their ubiquitous horizontal transfers.
In addition, although DNA transposons could be used as genetic engineering tools for insertional mutagenesis or transgenic vectors in previous applications, only a few of them have been widely used, such as Sleeping Beauty (SB).
The researchers explored new transposons with different functional properties. Among them, the most active transposon, Mariner2_AG (MAG), largely outperformed widely used vectors in CAR-T cell therapy, including lentivirus and SB, indicating its potential for clinical application.
The newly identified transposons with diverse functional characteristics described in the current study significantly expand the genetic engineering toolbox based on DNA transposons, supporting various application scenarios.
Overall, by utilizing the abundant genetic resources of the animal kingdom, the researchers conducted the largest-scale DNA transposon activity screening ever undertaken, resulting in the largest active DNA transposon dataset to date.
This study not only highlights the diverse functional and evolutionary characteristics of DNA transposons, but also expands the genomic engineering toolbox.
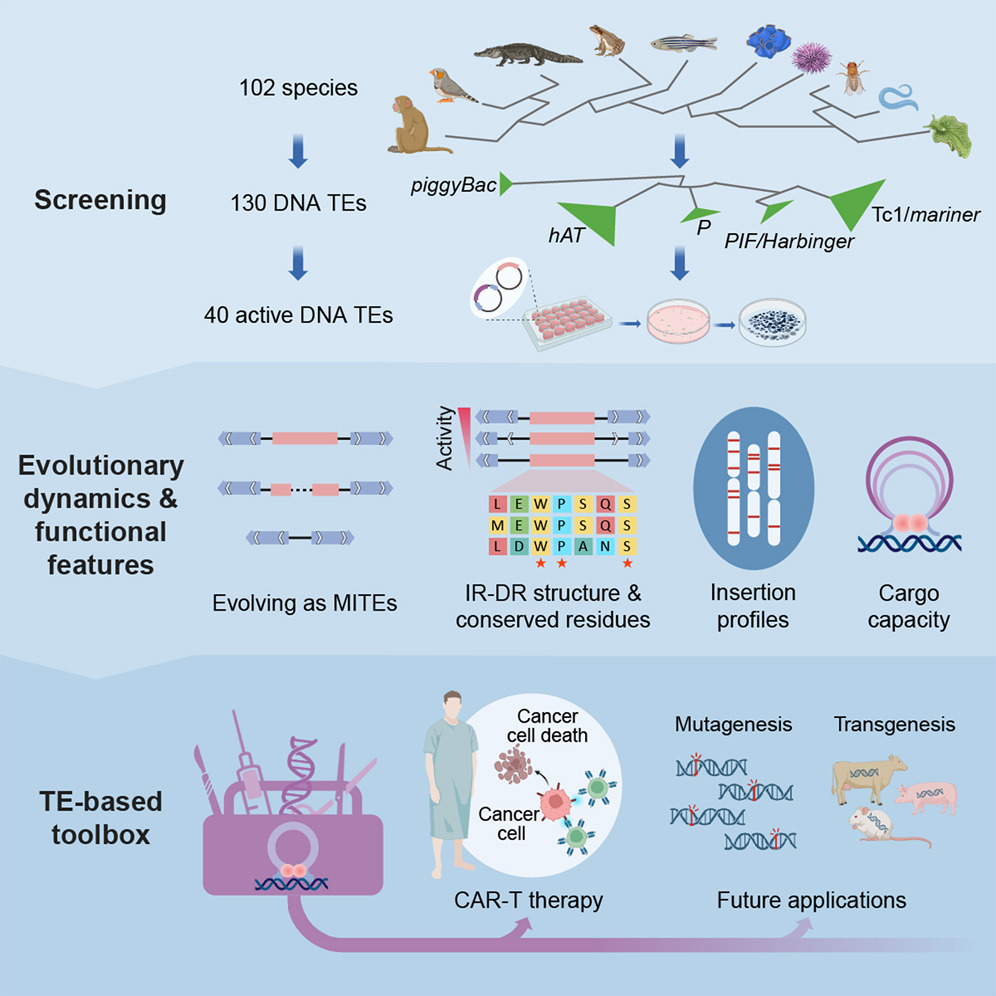
The comprehensive process of screening, analyzing, and applying active DNA transposons (Image by ZHANG et al.)

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

