Ethylene is a versatile gaseous plant hormone that plays an important role in rice growth, development, and environmental adaptation. Translational control allows the mRNA to be translated in an mRNA-selective, signal-responsive, and rapid manner. Previous studies in Arabidopsis have shown that ethylene can induce EIN2-mediated specific translational suppression of EBF1/2 mRNAs for activation of downstream signaling. However, whether any RNA-binding proteins are involved in this process remains unknown.
A research team led by Prof. ZHANG Jinsong at the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences provided insights into the mechanism of translational regulation in rice ethylene signaling. They developed an efficient screening system and identified a series of rice mutants, mao huzi (mhz), a Chinese name for cat whiskers, to describe the scattered adventitious roots in ethylene response mutants.
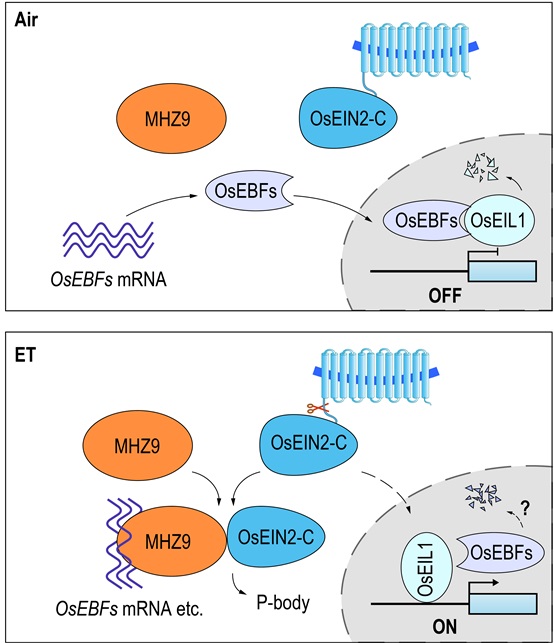
By analyzing of a rice ethylene response mutant, mhz9, they identified a glycine-tyrosine-phenylalanine (GYF) domain protein, MHZ9, which positively regulates ethylene signaling at the translational level in rice.
They found that MHZ9 is localizes to the RNA processing body (P-body). Biochemical studies further revealed that the C-terminus and N-terminus of MHZ9 play different roles. The MHZ9 C-terminus contains a glutamine-rich segment that mediates the interaction between MHZ9 and OsEIN2-C and is also sufficient and necessary for the P-body localization of MHZ9. The MHZ9 N-terminus contains a predicted domain for RNA splicing/modification, and binds directly to OsEBF1/2 and many other mRNAs.
Using multiple biochemical methods, they found that MHZ9 targets OsEBF1/2 mRNA for translational inhibition upon ethylene treatment. Suppression of OsEBFs translation would reduce OsEBFs-mediated degradation of the transcription factor OsEIL1, allowing OsEIL1 to accumulate for further activation of downstream ethylene signaling events.
Then, they performed ribo-seq analysis to investigate the role of MHZ9 in translational regulation at the whole-genome level during the ethylene response. Ribo-seq analysis indicates that 1956 genes would be significantly and differentially regulated at the translation efficiency (TE) level in response to ethylene. MHZ9 mediates the translational response of 1788 (~91%) of TE-altered genes induced by ethylene directly or indirectly.
They also observed multiple phenotypic changes in the mhz9 mutant under field-grown conditions, including dwarf, tiller number decrease, tiller spreading, and grain size reduction, suggesting that MHZ9 may have other roles in the control of agronomic traits in rice.
This study identifies a master regulator, MHZ9, for translational control during the ethylene response in rice. MHZ9-mediated translational regulation may facilitate the improvement of agronomic traits and adaptation to adverse environments for crops.
A proposed working model for MHZ9-mediated ethylene signaling in rice. In the absence of ethylene, MHZ9 remains at low-affinity-binding states to OsEBFs mRNA. OsEBFs proteins accumulate and lead to the degradation of OsEIL1. Ethylene response is OFF. With ethylene, MHZ9 possibly receives a signal from OsEIN2-C through direct interaction, and then binds to the 3’UTR of OsEBFs mRNA and other targets for translational inhibition, which contributes to the OsEIL1 protein accumulation and downstream ethylene signaling. The residual OsEBF1/2 may decay through unknown mechanisms. (Image by IGDB)