High-order chromatin structure is a prerequisite for the function of cis-regulatory elements in the genome, which plays an important role in gene regulation. In eukaryotes, the organization of three-dimensional (3D) genome presents a hierarchical pattern, where chromatin can be divided into different structural domains, such as chromosome territory, A/B compartment, topologically associated domain (TAD) and chromatin loop. There have been many studies about the dynamic changes of 3D genome during embryonic development on mammals. However, in-depth studies on genetic diversity of 3D genome have not been carried out in plants, especially in higher plants. And the relationship between genomic variation and 3D genomic variation, the effect of 3D genomic variation on crop domestication are still poorly understood.
A research group led by Prof. TIAN Zhixi from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences has investigated the genetic diversity and constructed a pan-3D genome of soybean, revealing the internal relationships among soybean genome, 3D genome and gene expression. The study was published in
Genome Biology.
The researchers obtained high-quality 3D genome data by performing high-throughput chromatin conformation capture experiments of 27 soybean germplasm materials that were de novo assembled in a previous study.
To investigate the conservation and variability of the 3D genome, they performed pan-omics analysis and constructed the pan-3D genome of soybean. According to the researchers, A/B compartments were generally conserved among soybean accessions, and the variation of A/B compartments was closely related to genomic features. Furthermore, regions with intermediate genomic features were the main region of A/B compartment switching.
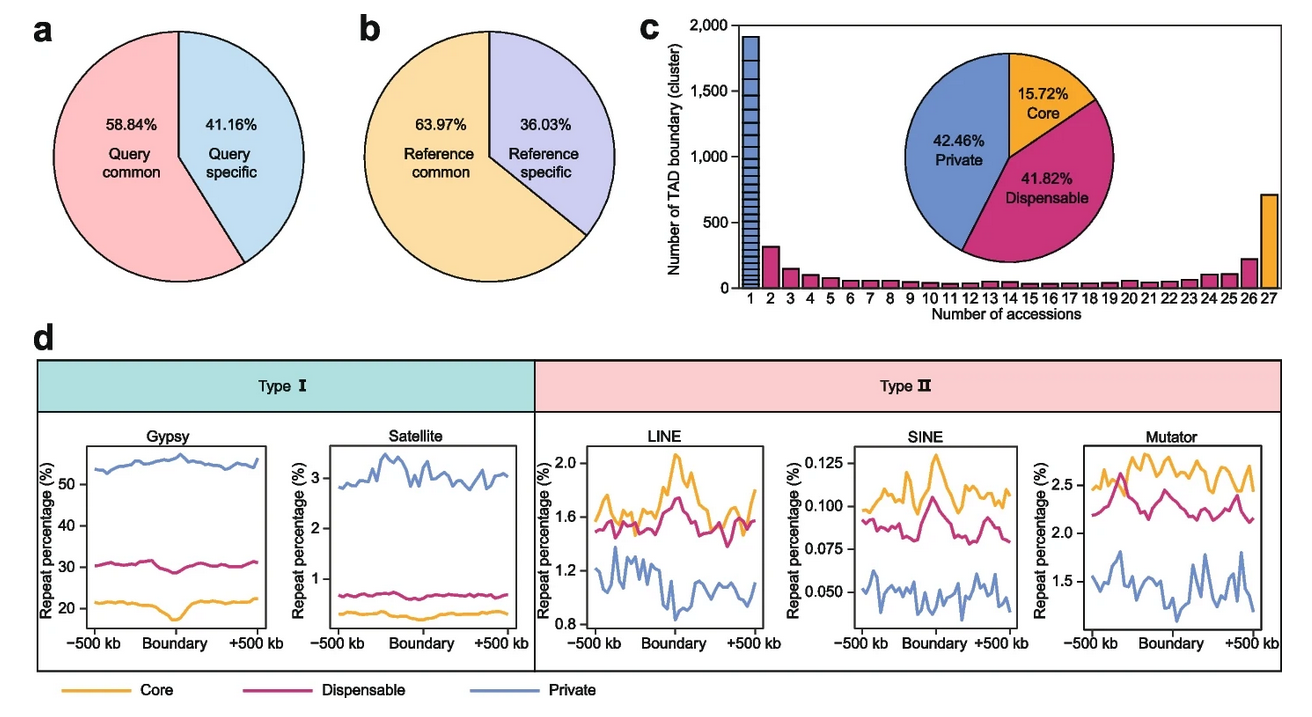
TAD boundaries define the range of interactions of TADs. In many cases, the variation of TAD boundaries represents a change in gene regulation. The researchers also constructed a pan-3D genome of TAD boundaries. Pan-3D genome showed that TAD boundaries had a higher level of variation than A/B compartments. Further analysis revealed that non-LTR retrotransposons (LINE and SINE) were enriched around TAD boundaries, suggesting that these two types of elements have important functions in maintaining TAD boundaries.
Besides, Gypsy elements and satellite repeats were enriched around accession-specific TAD boundaries, indicating that these elements play unique roles in the formation of specific TAD boundaries. These results explain how transposable element (TE) superfamilies reshape the 3D genome in plants for the first time.
Genomic structural variations (SVs) are the main source of genetic variation. Due to the lack of high-quality SV data, the relationship between 3D genomic variation and genomic SV has not been investigated in plants. The researchers further explored the relationship between genomic SV and 3D genomic variation based on high-quality SV data from de novo genome assembly. The research showed that Presence and Absence Variation plays the most important role in 3D genomic variation. Further analysis showed that the content of Gypsy elements and satellite repeats was significantly increased in the SVs that formed the accession-specific TAD boundaries.
These results confirmed that TEs can reshape the evolution of 3D genome by driving SVs. To explore the relationship between 3D genome diversity and gene expression, they verified the correlation between 3D genome and gene expression at multiple levels.
In addition, they also explored the selection process of 3D genome in wild soybean, landraces and cultivars during domestication and improvement. They found that the selection of 3D genome mainly occurred during domestication, rather than during improvement. This selection reshaped the gene regulation and led to changes in soybean gene expression.
"This work investigates the genetic diversity of 3D genome among plant germplasm through pan-3D genome construction, reveals the roles of plant TEs in reshaping 3D genome, analyzes the 3D genome variation caused by genomic SVs, and the selection and subsequent functional effects of 3D genome during crop domestication. These studies provide a new way to understand the evolution of plant genome, and also provide valuable resources for molecular design breeding," said TIAN Zhixi, the group leader and corresponding author of the study.
Pan-3D genome of TAD boundaries and TE distribution patterns. (Image by NI et al.)