
A recent study published in Nature Biotechnology revealed the development of high-fidelity Cas13 variants with markedly reduced collateral effects by mutagenesis and demonstrated the feasibility of hfCas13 for efficient on-target RNA degradation with almost no collateral damage in mammalian cells and animals.
This work was performed by researchers in Dr. YANG Hui’s Lab at the Institute of Neuroscience, Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, State Key Laboratory of Neuroscience and R&D team from HuiGene Therapeutics Co., Ltd.
The CRISPR-Cas13 system is a highly efficient tool for programmable RNA targeting, and has been harnessed for nucleic acid detection and RNA manipulations in various cell types and organisms. However, concerns have been raised over the off-targeting effects of Cas13. Previous structural studies have implicated that upon binding of Cas13 to target RNA, the two HEPN nuclease domains could form a catalytic site on the protein surface, providing potential for promiscuous cleavage of bystander RNAs apart from the on-target cleavage of target RNAs.
Due to this so-called collateral effect, Cas13 could degrade both target and non-target RNAs at random, thus making it difficult to design experiments and interpret results when using Cas13.
Therefore, attempts to diminish or eliminate promiscuous RNA degradation via engineering approaches are required for basic research and future in vivo applications of Cas13 system.
The researchers designed a dual-fluorescent reporter (EGFP & mCherry) plasmid system to detect the collateral effects of Cas13 in mammalian cells. In this system, loss of mCherry fluorescent cells is interpreted as successful on-target editing; loss of EGFP fluorescent cells is considered to represent off-target cleavage. Results showed that either Cas13a or Cas13d could induce significant collateral effects.
Then the researchers sought to engineer Cas13d (RfxCas13d or CasRx, Cas13d from Ruminococcus flavefaciens XPD3002) via mutagenesis and screen for variants with minimal collateral effects, based on a new well-designed dual-fluorescent reporter system comprised of a single plasmid containing EGFP, mCherry, and EGFP-targeting gRNA, together with each Cas13 variant.
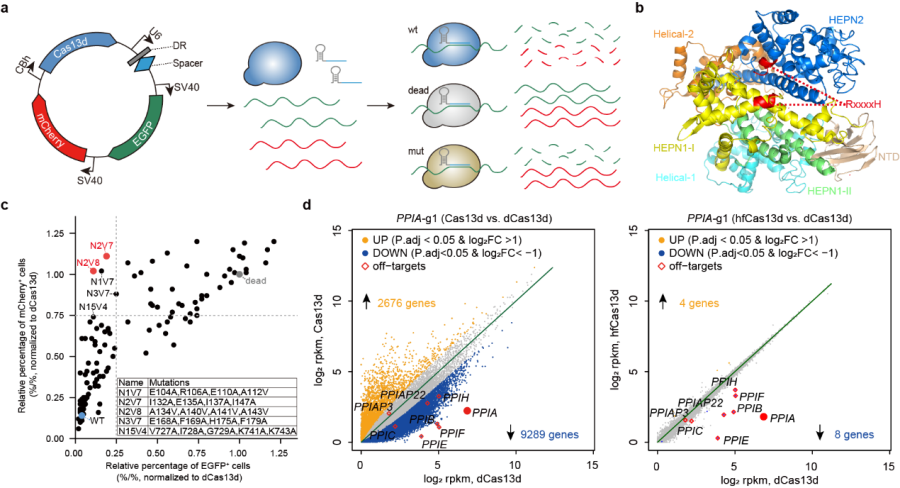
Fig 1. Design, screening, and characterization of engineered high-fidelity Cas13 variants (Image by CEBSIT)
YANG’s team designed and generated a mutagenesis library of Cas13d variants, and individually transfected them into HEK293 cells. By analyzing reporter fluorescence using flow cytometry, five variants (N1V7, N2V7, N2V8, N3V7, and N15V4) exhibited relatively low percentage of EGFP+ cells but high percentage of mCherry+ cells, indicating high on-target activity with reduced collateral activity.
The variant N2V8, termed as high-fidelity Cas13d (hfCas13d) for its highest specificity for RNA degradation, was used for further characterization including transcriptome-wide off-targets analysis using RNA sequencing. The hfCas13d showed marked reduction in the number of off-target genes when targeting several endogenous transcripts, such as PPIA. Moreover, hfCas13 variants showed no detectable collateral damage in cell lines, transgenic animals, and somatic cells, supporting their in vivo applications.

Fig 2. Cellular and in vivo consequences of collateral effects by wild-type Cas13d and their elimination using hfCas13d. (Image by CEBSIT)
Importantly, YANG and his team identified CRISPR-Cas13X system in 2021, the smallest (only 775 amino acids) RNA editing tool at present. In their new study, the researchers further engineered Cas13X and obtained hfCas13X variant exhibiting high specificity of on-target RNA degradation but minimal collateral effects. hfCas13X will show great application potential in the field of gene therapy based on RNA editing.
This work entitled “High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects” was published online in Nature Biotechnology on Aug 11, 2022. TONG Huawei, HUANG Jia, XIAO Qingquan, HE Bingbing, DONG Xue and LIU Yuanhua are co-first authors with equal contribution.

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

