
Cytidine-to-Uridine (C-to-U) RNA editing is a prevalent nucleotide modification at the RNA level in higher plant organelles, which has important roles in altering genomic genetic information and restoring amino acid conservation. Previous studies have revealed that RNA editing is specific in terms of tissues, suggesting that RNA editing may function as a regulatory device in plastid gene expression. However, to date, little is known about the dynamic distribution of RNA editing among tissues in plants.
Researchers from the Wuhan Botanical Garden of the Chinese Academy of Sciences performed a bioinformatics analysis of publicly available RNA-sequencing data from four tobacco tissues (root, stem, leaf, and flower) to explore the dynamic landscape of RNA editing between tissues.
According to the researchers, the distribution of editing sites was uneven among tobacco tissues. Compared with the other three tissues, the root tissue had the least editing sites and the lowest average editing efficiency.
The expression analysis of 60 RNA edited genes displayed that most of the RNA edited genes (54 out of 60) were expressed in all tissues with no bias, while the overall expression level was highest in stem, but lowest in root.
Moreover, they found that the 11 tissue-specific edited genes almost expressed in all tissues at a low level except rrn26 (rRNA 26) and rps14 (Ribosomal protein S14). For some tissue-specific genes, they were expressed at lower levels in a particular tissue but underwent editing only in that tissue.
In fact, there was no causality between expression and editing of transcripts in this study, and it seems that different types of tissues may generate different kinds of signals to affect RNA editing and expression of organelles genes separately.
RNA editing factors, like PPR (Pentatricopeptide repeat) and MORF (Multiple organelle RNA editing factor) proteins, also showed tissue-specific expression, especially morf genes, which may partly explain the reason for the varied RNA editing events among different tissues.
Besides, due to the complexity and diversity of plant RNA editosome, the regulatory relationship between RNA editing factors and editing sites is not simply one-to-one, so further studies are needed to explore the targeting relationship between them.
Results were published in Plant Cell, Tissue and Organ Culture entitled "Dynamic landscape of mitochondrial CytidinetoUridine RNA editing in tobacco (Nicotiana tabacum) shows its tissue specificity."
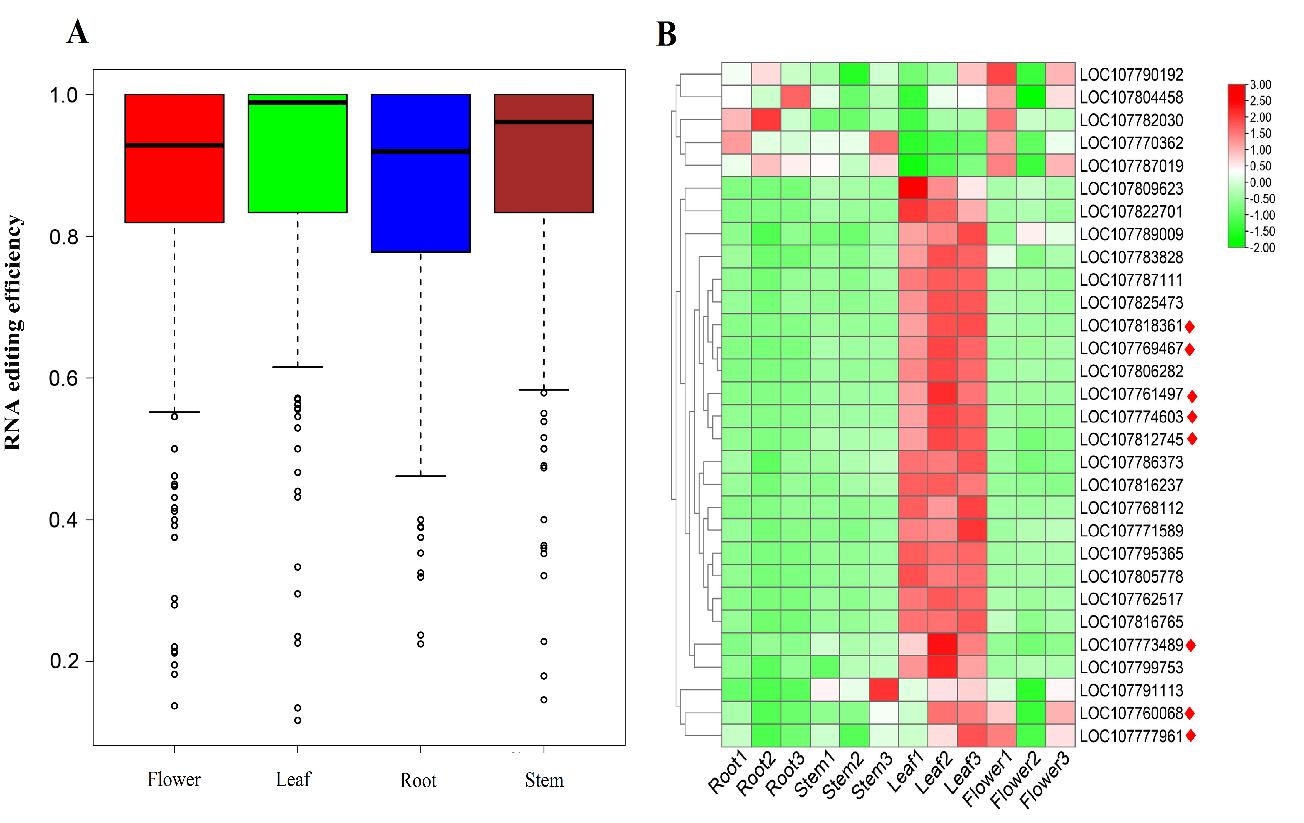
(A) Overall average RNA editing efficiency in four tissues of Nicotiana tabacum mitochondrial genome. (B) Heatmap of RNA editing factors expressions in four tissues of tobacco mitochondrion (Image by WBG)

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

