
RNA splicing is a critical step to generate mature RNA, and alternative splicing (AS) is the primary mechanism for mammalian cells to diversify their proteomes. Aberrant splicing impairs many biological processes and has been implicated in an increasing number of diseases.
In contrast to gene expression that has many efficient platforms to manipulate (e.g.RNAi, CRISPRi, or CRISPR-mediated gene disruption), splicing has been difficult to modulate especially in its native context.
Development of an efficient genetic approach to modulate splicing is also pertinent to the treatment of many severe hereditary diseases caused by aberrant splicing. Antisense Oligonucleotides (ASOs) have been exploited to correct splicing defects in some genetic diseases, such as Duchenne muscular dystrophy (DMD), affecting one in 5,000 males at birth.
While ASO therapies only have the moderate effect (e.g. 1 of the 12 patients had increased dystrophin expression), they require expensive chemical modification and life-long treatment, adding enormous economic burdens (>$300,000 per year) on patients and society. Thus single-dose cures for these diseases are still in dire need.
In a recent study led by Dr. CHANG Xing from Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences, researchers developed the first genetic approach to efficiently modulate various types of RNA splicing in their native chromatin context.
Based on the CRISPR-guided cytidine deaminase (i.e. Targeted AID-mediated mutagenesis, TAM), this approach precisely edits universal cis-regulatory elements of splicing to either suppress or promote inclusion of an exon, enabling both loss-of-function and gain-of-function analysis using the same platform, namely, to promote cassette exon skipping or inclusion, to induce switch between mutually-exclusive exons, to retain very short introns, and to enhance the usage of alternative splice sites.
Moreover, researchers further demonstrated that this approach is able to efficiently correct splicing defects identified in human diseases. Applying TAM, they have successfully restored reading frame and dystrophin expression of a mutant DMD gene (encodes dystrophin protein) in iPSCs derived from a patient with Duchene muscular dystrophy.
This study demonstrates splicing modulation via TAM fulfills a long-term, unmet, but pressing need to understand the functions of splicing isoforms and is of great therapeutic value to the treatment of many diseases, including DMD.
This work has been published online ahead of printing in Molecular Cell on Oct 4th, 2018 tilted "Genetic modulation of RNA splicing with a CRISPR-guided cytidine deaminase".
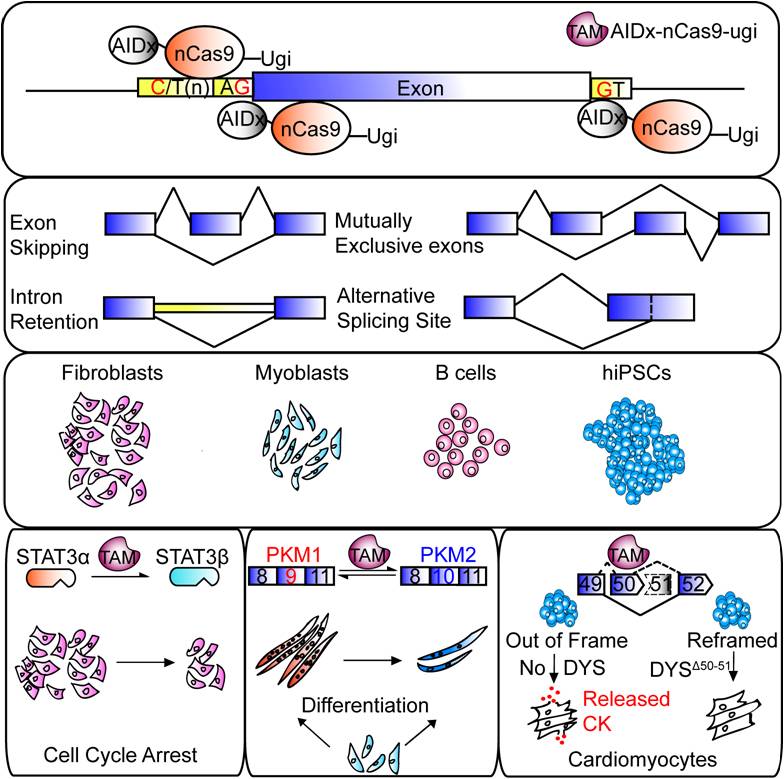
TAM (AIDx-nCas9-Ugi fusion protein) modulates RNA splicing and corrects splicing defects in human diseases (Image by Dr. CHANG Xing's Group)

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

