
An international group of researchers, led by Professor ZHU Yongguan from the Institute of Urban Environment, Chinese Academy of Sciences, have detected the widespread presence of antibiotic resistance genes in estuarine areas - regions where land, freshwater and seawater meet.
The results indicate that polluting antibiotic agents and resistance genes, released into the natural environment by human activity, are causing potentially dangerous changes to local bacteria in estuaries.
Antibiotics are chemical reagents that frequently used to treat infections caused by bacteria, parasite, and fungi. The discovery and introduction of antibiotics in medicine to treat infections have dramatically changed human health.
Bacteria that can't be effectively killed by antibiotics are known as antibiotic resistant. Antibiotic resistance is often conferred by a specific gene, these genes are called antibiotic resistance genes (ARGs).
The emergence and spread of antibiotic resistance is an increasing threat to human health due to antibiotics treatment failure, leading to increasing cost for health care and even infectious death.
Antibiotic resistance has been well documented in clinical settings, however, there are global concerns on the spread and dissemination of antibiotic resistance in the environments.
The study, which tested 4,000 km of China’s coastline, found that estuaries generally had levels of antibiotic resistance gene pollution at around one million resistance genes per gram of estuarine sediment, and in some cases up to 100 million per gram.
This study tells us that antibiotic resistance genes are readily found in bacteria in estuaries - the places tested in China are just a microcosm of a worldwide problem.
In this study, authors detected over 200 different resistance genes, 18 of which were found in all the sediment samples tested, meaning that there are quite a variety of antibiotic resistance genes that could help estuary bacteria along the path to becoming superbugs.
They estimated that on average one bacterial cell contain roughly one antibiotic resistance gene-meaning that the great majority of bacteria had at least one gene that helped them become resistant to antibiotics.
Despite the large spatial scale of sampling, they found no geographical patterns, and further analysis showed that the major cause of the accumulation of these genes in the environment is human activity, and the failure of waste water treatment plants to remove these genes during processing - something that we need to focus on achieving in the future. Otherwise, we could find ourselves facing dire environmental, agricultural and medical consequences.
These findings were recently published in Nature Microbiology entitled "Continental-scale pollution of estuaries with antibiotic resistance genes".
This study was conducted with collaborators from University of Hong Kong, Tsinghua University and Macquarie University.
This study was financially supported by the National Key Research and Development Plan, the Natural Science Foundation of China, the Knowledge Innovation Program of the Chinese Academy of Sciences, the International Science & Technology Cooperation Program of China and Youth Innovation Promotion Association CAS.
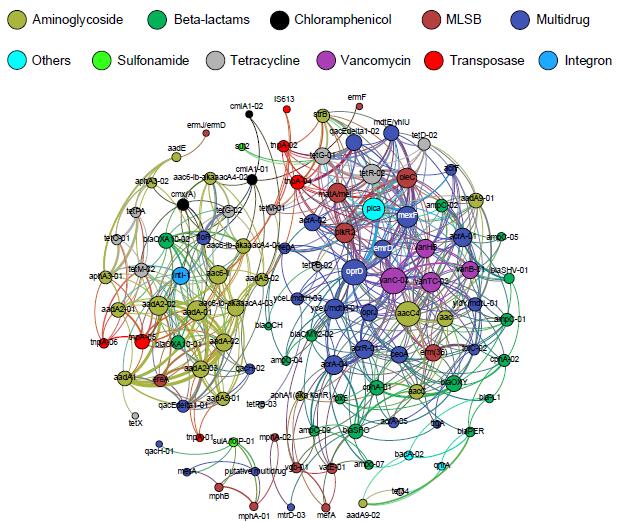
Network analysis depicting co-occurrence patterns amongst antibiotic resistance genes, transposases and the class 1 integron-integrase gene (intI1). The significant cluster of resistance genes around intI1 (P < 0.01) consists of known gene cassettes associated with clinical class 1 integrons. (Image by ZHAO Yi)

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

