
Drug discovery and development is notoriously difficult despite the plethora of advanced experimental and computational technologies being put to task. Surging dataset sizes, poorly understood biological mechanisms of action, computational workflows bedeviled by mixes of large and small files are all part of the challenge.
Increasingly HPC is playing an important role to relieve the bottleneck (see HPCwire article, 25 Percent of Life Scientists Will Require HPC in 2015). A new paper by prominent researchers in China – ‘Applying high performance computing in drug discovery and molecular simulation’ – examines China’s growing use of HPC in pursuit of new drugs. The paper was published online in this month’s issue of National Science Review, by Oxford Journals.
The work is less a deep dive into the life science and computational challenges and performance metrics than a sweeping survey of the drug discovery applications to which HPC is being applied in China and a review of some of the large computational resources being used. For example, Tianhe-2, the top performer in Top500, has been used in drug repositioning research.
"...Computational drug discovery and design (CDDD), which is based on HPC…has become a critical approach in drug research and development and is financially supported by the Chinese government. This approach has yielded a series of new algorithms in drug design, as well as new software and databases. This review mainly focuses on the application of HPC to the fields of drug discovery and molecular simulation at the Chinese Academy of Sciences," write the authors affiliated with State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences.
Shown below are MD simulations run on high-end HGPC resources taken from a figure in the paper.
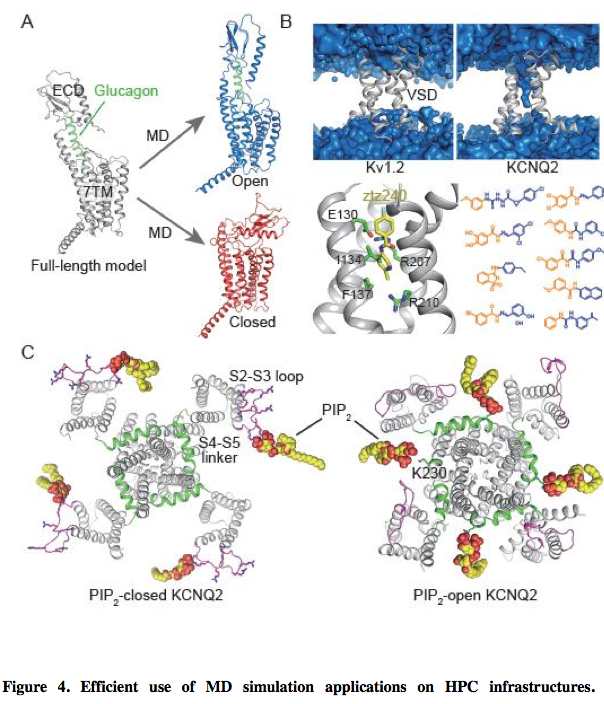
(A) Identification of open (blue) and closed (red) states of full-length GCGR in MD simulations of full-length GCGR model (grey). The structures of full-length GCGR and glucagon (green) are shown as cartoons and glucagon is semi-transparent. (B) An activator-binding pocket was discovered in the VSD of the KCNQ2 potassium channel and nine activators were identified through structure-based virtual screening.
The surface of water molecules in MD simulations of Kv1.2 and KCNQ2 channels are shown in blue, with VSDs in grey cartoons. Residues involved in interactions with ztz240 in the binding model of ztz240 in the VSD of KCNQ2 are displayed as green sticks. Ztz240 is depicted as yellow sticks. The fragments orientated toward the intracellular end of the VSD in the chemical structures of nine identified activators are highlighted in yellow, while the other ends are highlighted in blue. (C) PIP2 interactions with the closed and open states of the KCNQ2 channel in MD simulations. PIP2 molecules are depicted as yellow spheres. S2-S3 loops and S4-S5 linkers in KCNQ2 are respectively highlighted in magenta and green. Residues contributing to the stable binding of PIP2 are shown as sticks, such as positively charged residues in the S2-S3 loop and K230 in the S4-S5 linker.
The authors thanked the National Supercomputing Center in Jinan, the National Supercomputing Center in Tianjin (Tianhe-1A) and the Tianhe Research and Development Team of the National University of Defense Technology (Tianhe-2), in making the studies possible. “Because the biological systems studied using all-atom MD simulations can be very large, comprising millions of atoms, the calculations required are often too complex and computationally intensive for even the best supercomputers. It is clear that there is room for improvement. Steady growth in HPC has made MD simulation spans wider spatial and temporal ranges and resolutions, which are likely to play an increasingly important role in the development of novel pharmacological therapeutics,” write the authors. (HPC Wire)

86-10-68597521 (day)
86-10-68597289 (night)

86-10-68511095 (day)
86-10-68512458 (night)

cas_en@cas.cn

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

