
Newsroom
Saccharina japonica, one of the most widely cultivated seaweeds globally, is particularly prominent along China's coastline, where it has been farmed in both cold-temperate and subtropical waters. Although traditional selective breeding and hybridization have yielded several high-quality varieties, the absence of a complete genome assembly has hindered further genetic advancements.
The primary challenge in sequencing the kelp genome lies in DNA extraction. Due to its high polysaccharide content, including alginate, DNA purification is notoriously difficult, which compromises sequencing quality and assembly accuracy. Previous genome assemblies were highly fragmented due to technological limitations, hindering progress in molecular breeding and genomics research.
To address these challenges, a research team led by Prof. PANG Shaojun from the Institute of Oceanology of the Chinese Academy of Sciences (IOCAS) developed a novel sample preparation method. Using parthenogenetic techniques, the team cultivated a homozygous diploid female sporophyte for genome sequencing. This approach minimized heterogeneity, significantly enhancing the accuracy and reliability of the genome assembly.
This study was published in Scientific Data.
The researchers achieved a high-quality, chromosome-level genome assembly of S. japonica by combining PacBio HiFi, Illumina short reads, and Hi-C data. The final genome size is 516.11 Mb, with 96.15% of the sequence anchored to 32 chromosomes. This assembly is not only more complete but also exhibits continuity compared to previous versions.
This progress provides a critical resource for molecular breeding and genetic engineering in kelp, enabling improved cultivation techniques and contributing to sustainable aquaculture practices.
"With over 17,000 protein-coding genes identified and functional annotations available for the majority, our work paves the way for future ecological and evolutionary studies of brown algae," said Dr. LI Xiaodong, the study's first author.
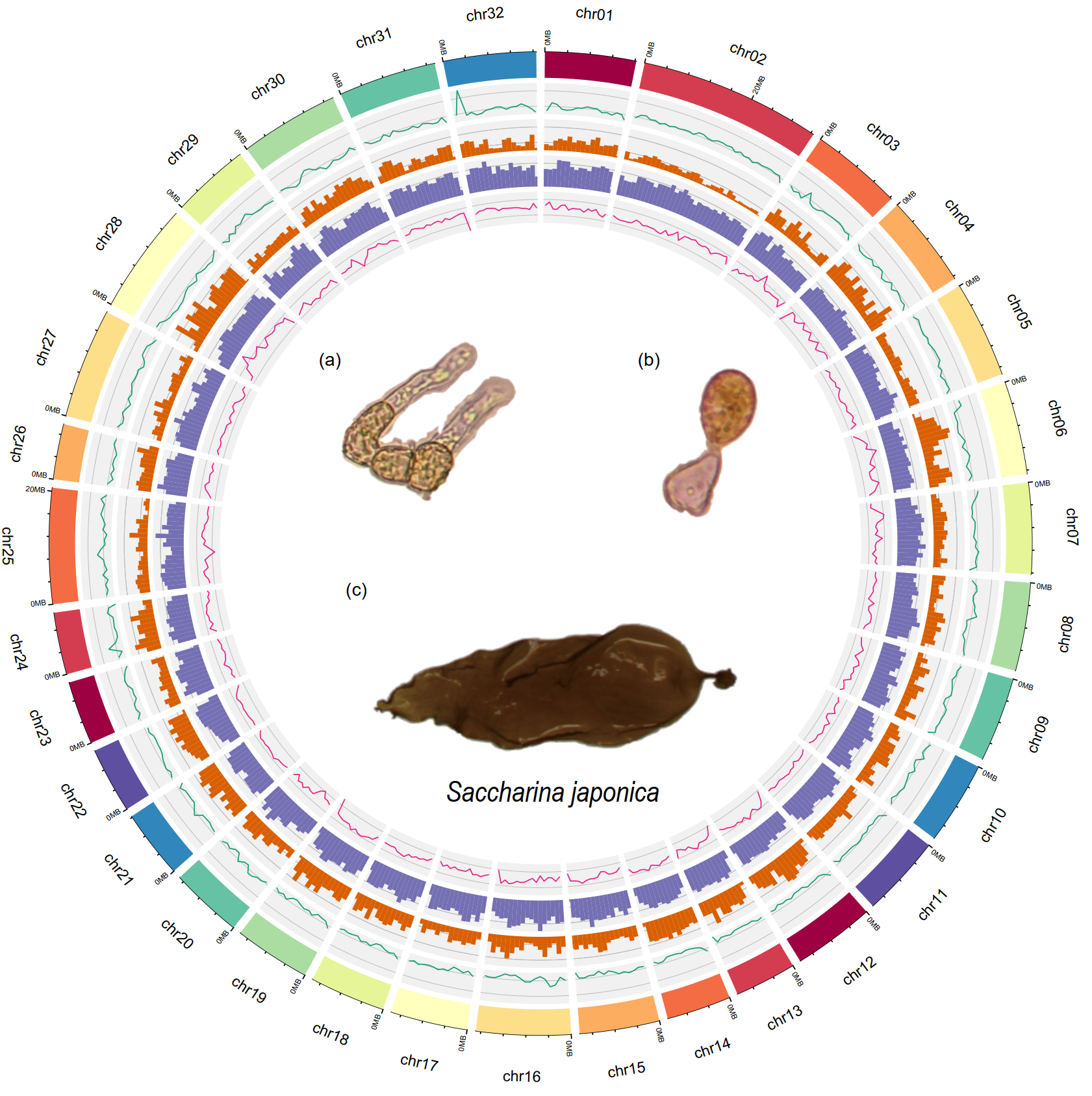
Fig. 1 Chromosome-level genome assembly of S. japonica. (A) Circos plot illustrating the chromosome structure, short-read depth, gene density, repetitive element density, and GC content. The visual highlights the genome's organization from the outermost to the innermost layers, with sections corresponding to the female gametophyte (a, b) and female sporophyte (c).
