
With a consistent global annual production of more than 100 million tons, the sweet potato, Ipomoea batatas, is an important source of calories, proteins, vitamins and minerals for humanity. It is the seventh most important crop in the world and the fourth most significant crop of China.
An international team of researchers from the Chenshan Botanical Garden in Shanghai, the Max Planck Institute for Molecular Genetics in Berlin, the Shanghai Institute of Plant Physiology and Ecology (SIPPE), the Max Planck Institute of Molecular Plant Physiology in Potsdam, the Tai’an Academy of Agricultural Sciences in Shandong, has been able to sequence and phase the sweet potato genome. The study, published in Nature Plants, shed light on the evolutionary history of the sweet potato.
Sweet potato has the most complicated genome among flowering plants in the Convolvulaceae family, being hexaploid and having 90 chromosomes (2n=6x=90). With the novel haplotyping method developed in the study, scientists demonstrated that 30 chromosomes came from the diploid progenitor of sweet potato and the other 60 chromosomes came from the tetraploid progenitor, and modern cultivated sweet potato originated from a crossing between the diploid and tetraploid progenitors about 500,000 years ago following a whole genome duplication event.
There are 78,781 gene models from 49,063 gene loci in current assembly. The study showed that, due to functional redundancy among the six homologous chromosomes in the hexaploid organism, quite a number of the genes have deleterious mutations in different alleles, such as a high frequency of frameshift events identified in the phased six haplotypes. The result indicated that the natural selection pressure on particular genes in the hexaploid organism is much lower than those in diploid organisms, which also provides the basis for faster genome evolution in the polyploid organisms.
Also, many gene clusters were found in current genome assembly, implying that pathway regulation via clustered genes is commonly used in the genus Ipomoea. Although all the orthologous genes found are based on protein sequence similarity, their biological functions are not necessarily the same as those reported for similar gene clusters in other species. Nevertheless, the identified gene clusters in I. batatas open up possibilities for investigating metabolic regulatory mechanisms in this plant.
Overall, the methodology presented in the paper is an important advance in the genomic analysis of polyploid organisms. The results not only revealed the mysterious origination of hexaploid sweet potato, but also accelerated further utilization of wild relatives of sweet potato. Meanwhile, the haplotype-resolved genome sequence of sweet potato ushers the era of precise genome manipulation for this human food resource.
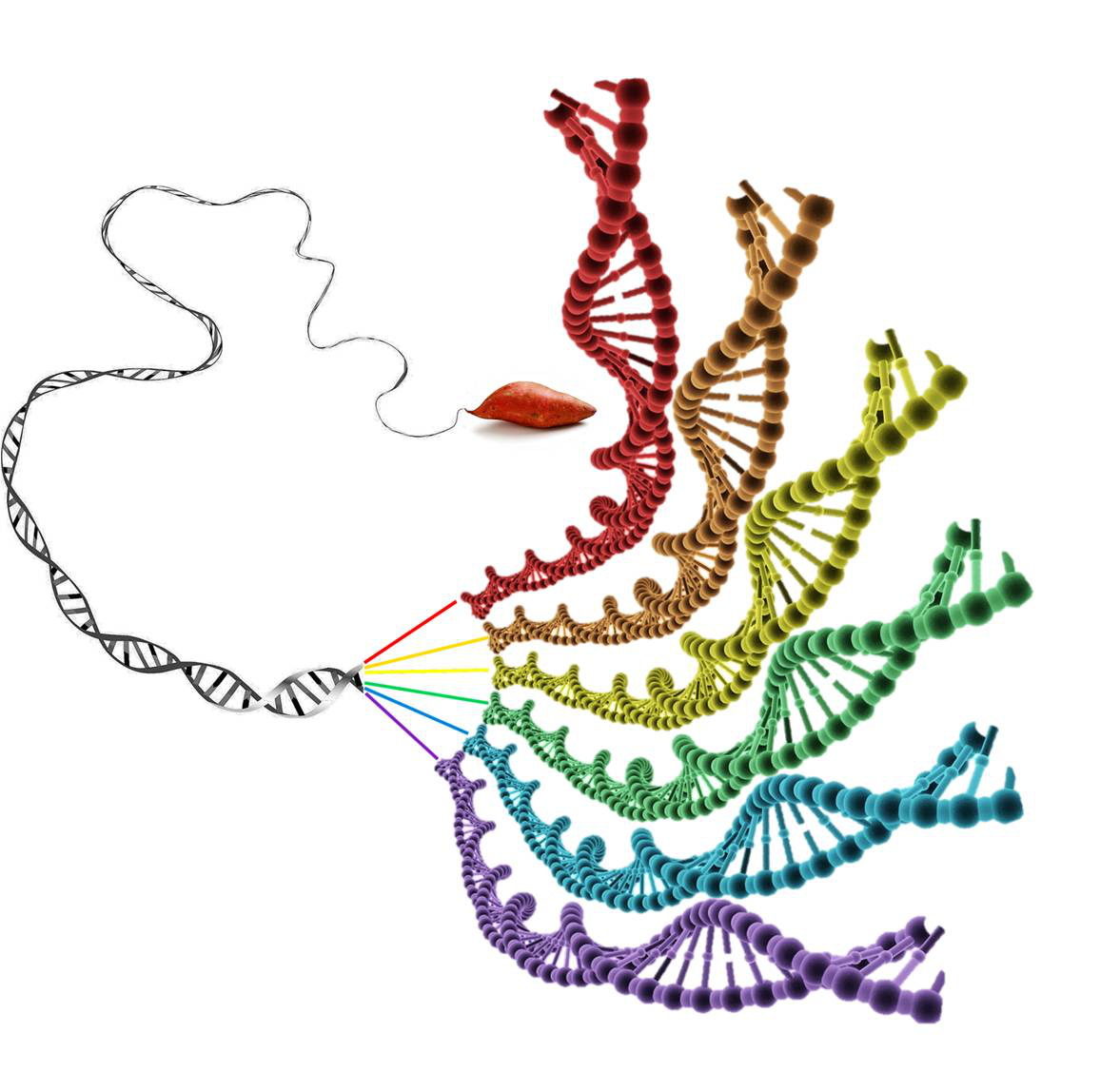
Figure: The haplotype-resolved genome sequence of hexaploid sweet potato (Image by Dr. ZHANG’s lab)

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

