
The Plos pathogens has published online the research progress on the replication mechanism of Hepatitis C virus (HCV) in cultured cells on April 30, 2010. This was a collaborative work done by Prof. Takaji Wakita’s group in NIH, Japan and Prof. Tetsuya Toyoda’s group in Institut Pasteur of Shanghai, CAS
HCV research is limited because only JFH1 can replicate and produce viruses efficiently in Huh7 cells. In order to find the mechanism of replication of JFH1, we firstly compared the structural and biochemical differences of RdRp between JFH1 and HCR6 (1b) and J6CF (2a), and identified the JFH1 RdRp specific amino acidswhich enhanced polymerase activity of HCR6 and J6CF. In the presence of the N3H region and 3’ untranslated region (UTR) of JFH-1, a combination of A450S, R517K and Y561F was sufficient to confer HCV genome replication activity and virus production ability to J6CF in cultured cells (Fig. 2).
Y561F was found to be encoded in the loop of 5BSL3.2 (CRE) and extended the annealing region for kissing-loop interaction of CRE and 3’XSL2 (Fig. 3). We next analyzed the 3’ structure of HCV genome RNA. Shorter polyU/UC tracts of JFH-1 were more favorable for efficient RNA replication than J6CF. Furthermore, 9458G in the JFH-1 variable region (VR) was found to be responsible for RNA replication activity by the suitable RNA structures.
In the current study, we investigated the relationships between HCV genome replication, virus production activity and the structure of N5BX. We found that N3H, high polymerase activity, enhanced kissing-loop interactions and optimal viral RNA structure of the 3’UTR were required for J6CF replication in cultured cells.
This work was supported bygrants from the Chinese Academy of Sciences, “973” and the Science and Technology Key Project for Infectious Diseases in the National Eleven Fifth Year Plan.
RNA Polymerase Activity and Specific RNA Structure are Required for Efficient HCV Replication in Cultured Cells
Asako Murayama1†, Leiyun Weng2†, Tomoko Date1, Daisuke Akazawa1,3, Xiao Tian 2, Tetsuro Suzuki1, Takanobu Kato1, Masashi Mizokami4, Takaji Wakita1* and Tetsuya Toyoda2*
1.Department of Virology II, National Institute of Infectious Diseases, Tokyo, Japan
2.Unit of Viral Genome Regulation, Institute Pasteur of Shanghai, Chinese Academy of Sciences, 225 South Chongqing Road, 200025 Shanghai, P. R. China
3.Pharmaceutical Research Lab, Toray Industries, Inc., Kanagawa, Japan
4.Department of Clinical Molecular Informative Medicine, Nagoya City University Graduate School of Medical Sciences, Nagoya, Japan
†These authors contributed equally
*Corresponding author
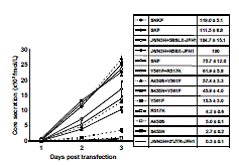
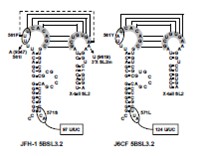
Fig. 2. Effect of A450S, S455N and R517K. Fig. 3. Comparison of 5BSL3.2and Y561Fmutations.

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

