
Newsroom
T-box riboswitches are an important class of gene regulatory elements whose function depends critically on their three-dimensional structure and conformational dynamics. However, the mechanism by which these riboswitches accurately recognize tRNAs with different aminoacylation states and subsequently initiate transcriptional or translational regulation remains unclear.
A research team from the Institute of Biophysics of the Chinese Academy of Sciences and Tsinghua University used single-molecule fluorescence resonance energy transfer (smFRET) to systematically investigate the structural dynamics of the Mycobacterium tuberculosis ileS T-box riboswitch.
The study uncovered the molecular mechanism by which the riboswitch discriminates the aminoacylation status of the tRNA 3′ end during co-transcription, thereby regulating the initiation of translation.
This work was published in Nature Communications on November 25.
The researchers selected multiple labeling pairs within the T-box aptamer and discriminator domains and combined UBP-based, site-specific fluorescent labeling with smFRET analysis to comprehensively monitor the structural transitions of the T-box riboswitch induced by the binding of tRNAs in different aminoacylation states.
They discovered that the T-box employs a conformational selection mechanism to achieve two-step recognition and identity verification, as well as aminoacylation-state sensing.
To further explore how co-transcriptional processes influence T-box regulatory mechanisms, the researchers generated T-box truncations of various lengths (to simulate distinct transcriptional intermediates) as well as fluorescently labeled co-transcriptional T-box/tRNA complexes, and performed additional smFRET measurements.
The results showed that recognition of the tRNA NCCA terminus by the T-box is heavily dependent on transcriptional progression. This co-transcriptional regulation enables the riboswitch to rapidly respond to intracellular amino acid availability, ensuring efficient and precise control of gene expression.
Based on these findings, the researchers proposed a co-transcriptional regulatory model for the T-box riboswitch, elucidating the complete dynamic process through which it sequentially recognizes tRNA, senses aminoacylation, undergoes conformational switching, and ultimately modulates gene expression during transcription.
This study provides important mechanistic insights into RNA-RNA interactions and lays a theoretical foundation for the development of new antibiotic strategies.
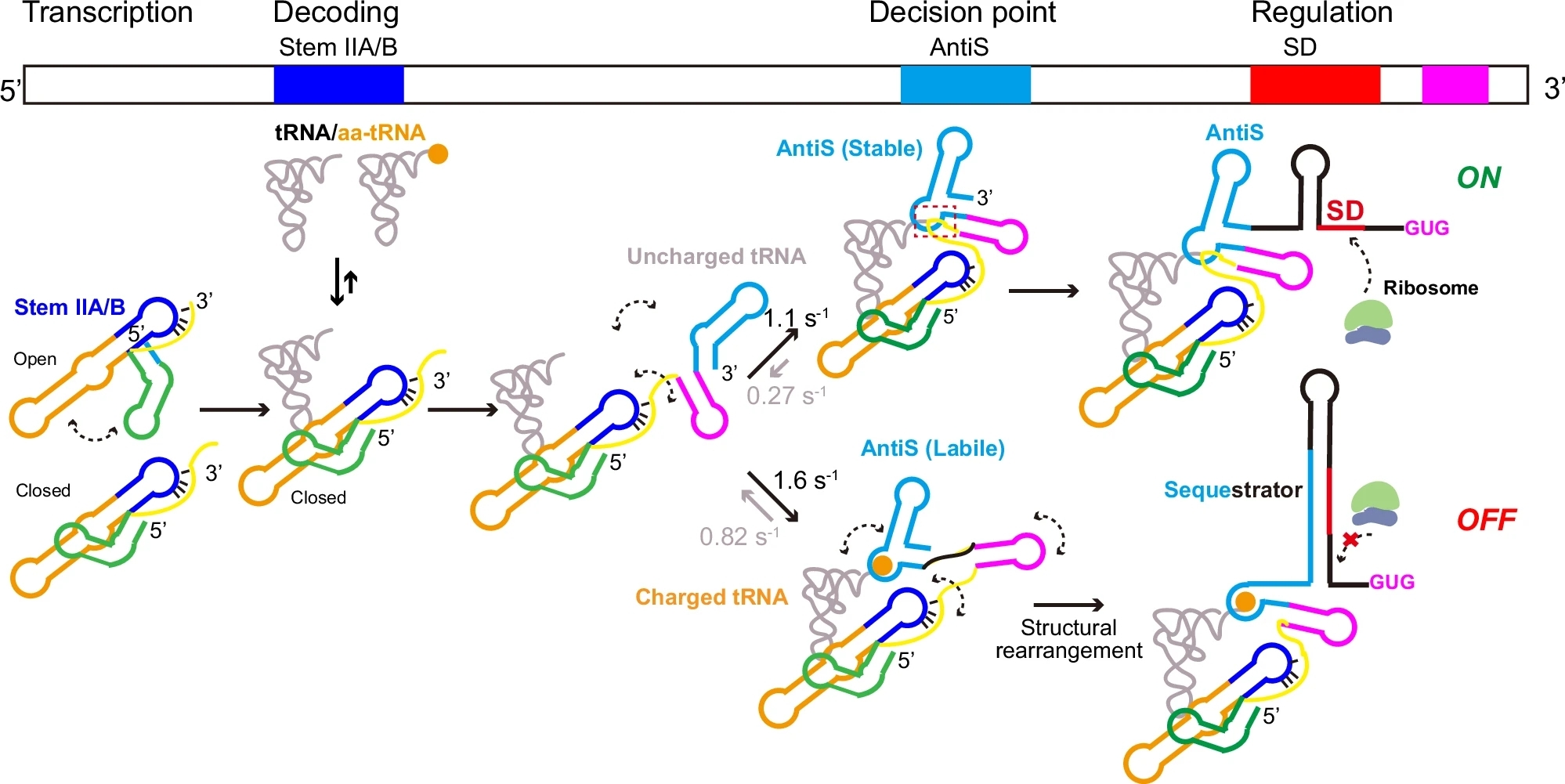
Model of Co-transcriptional Gene Regulation by the T-box Riboswitch (Image by FANG Xianyang's group)
