
Newsroom
Scientists from the Institute of Zoology of the Chinese Academy of Sciences (CAS) and CAS center for Excellence in Biotic Interactions have discovered that tobacco plants can recognize a potential elicitor protein cathepsin B3 (CathB3) in aphid saliva, and suppress aphid feeding by triggering reactive oxygen species (ROS) accumulation in phloem.
This novelty research was recently online published in Current Biology. The green peach aphid Myzus persicae, an extremely important insect pest of many crops worldwide, has been reported to colonize hundreds of plant species. M. persicae has a variety of host plant-associated lineages, which differ in fecundity and survival rate when reared on specific host plant species.
The tobacco-adapted (TA) M. persicae lineage performs well when feeding on tobacco plants, whereas non-tobacco-adapted (NTA) aphid lineages perform poorly. The molecular mechanisms underlying different host plant-associated lineages of M. persicae manipulate the chemical induced defenses of host plants are generally poorly understood.
"Since M. persicae lineages differ in their feeding efficiency and population fitness on tobacco plants, it is reasonable to speculate that some types of salivary component-triggered plant resistance could be lineage-specific," said SUN Yucheng, corresponding author of the study.
To do this, Prof. GE Feng, SUN Yucheng and their team screened differentially expressed genes using transcriptome analysis, to identify saliva-localized proteins by analysis of the salivary proteome of the TA vs. NTA M. persicae lineages.
They identified a conserved cysteine protease CathB3 that was constitutively up-regulated in the salivary glands of a natural NTA M. persicae lineage, when compared to a TA lineage.
More importantly, CathB3 of M. persicae could be secreted into host plants. When CathB3 was silenced, NTA aphids had quicker access to phloem and exhibited longer phloem feeding duration, which subsequently increased aphid growth rate and survival rate on tobacco plants.
The transient expression of the full-length CathB3 sequence in tobacco plants also confirmed that CathB3 could suppress the phloem feeding of TA aphids, suggesting that aphid CathB3 triggered an effective plant defense response against aphids, and attenuated the phloem feeding of NTA aphids.
CathB3 was found to specifically interact with the plant EDR1-like protein, a Raf-like MAPKKK, through its propeptide domain. NtEDR1-like was mainly expressed in vascular tissues.
To evaluate the function of EDR1-like in aphid feeding behavior, NtEDR1-RNAi lines with attenuated (irEDR1) and up-regulated (35S::EDR1) EDR1-like expression were generated.
Aphids that fed on 35S::EDR1 plants exhibited the shortest phloem feeding duration whereas aphids that fed on irEDR1 plants exhibited the longest phloem feeding duration, suggesting that EDR1-like mediated an effective defense of tobacco plant against aphids and reduced aphids' feeding efficiency. The RNA-seq analysis of tobacco plants revealed that EDR1-like protein was necessary for the activation of phloem ROS by plants.
During long-term co-evolution, some TA aphid lineages have developed improved phloem feeding efficiency, and have improved their fitness by releasing less CathB3 into plant tissue during feeding.
These findings reveal a novel function for a cathepsin-type protease in aphid saliva that elicits effective host plant defenses, and may be useful for the development of pest control strategies that are specific for piercing-sucking insects.
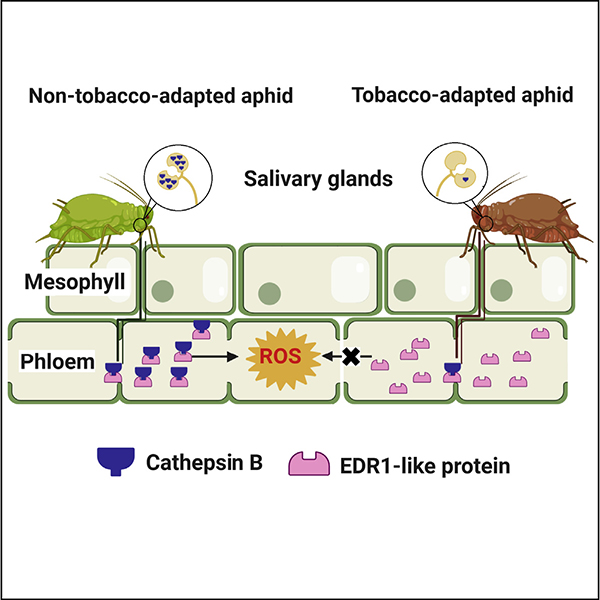
Figure 1. Two host associated aphid lineages triggered lineage-specific resistance of host plant (Image by IOZ)
