
Newsroom
CRISPR-Cas system is an adaptive immune system of bacteria and archaea to prevent against the foreign nucleic acids. Cas9 is an RNA-guided DNA endonuclease. Cas9 cleaves target dsDNA via its RuvC and HNH domains. In currently available DNA-bound, pre-cleavage Cas9 structures, the active site of the HNH domain is distant from the cleavage site of the target strand. The HNH domain is in the inactive state in all reported structures. To understand the mechanism of target strand cleavage, it is critical to elucidate the structures of DNA-bound Cas9 complexes in their catalytically competent states.
Two recently characterized Cas9 homologs from Neisseria meningitides have been developed as a compact and high-accuracy genome-editing tool. In 2016, three anti-CRISPR proteins (AcrIIC1-3) were reported to inhibit NmeCas9 by Karen Maxwell’s group from University of Toronto. AcrIIC2 was then found to prevent the assemble of the Cas9-sgRNA complex by WANG Yanli’s group from Institute of Biophysics of the Chinese Academy of Sciences and Karen Maxwell’s group. However, the complete molecular mechanisms for the DNA cleavage by NmeCas9s and the inhibition of the DNA cleavage by AcrIIC3 are still unknown.
In a study published in Molecular Cell, WANG’s group and Erik Sontheimer’s group from University of Massachusetts Medical School reported structures of the NmeCas9-sgRNA binary complex, a seed-matched, a fully paired pre-catalytic, a fully paired catalytically poised, and a post-TS-cleavage DNA-bound NmeCas9 ternary complexes, which represent five distinct states of NmeCas9 along its DNA cleavage pathway, showing the processes of DNA cleavage step by step as a movie.
These results provided a high-resolution view of the engagement of a Cas9 HNH domain with the guide-target heteroduplex, and showed important structural rearrangement of the REC2 and HNH domains of NmeCas9 during the target DNA binding and cleavage.
The HNH domain is flexible and exhibits a conformational equilibrium that usually favors positioning distant from the cleavage site. The researchers found that HNH mutants, which have higher binding affinity with the target, have enhanced DNA cleavage activity.
Besides, they also reported two structures of AcrIIC3-bound NmeCas9-sgRNA complexes with or without dsDNA. These structural studies revealed that two AcrIIC3 proteins tether two NmeCas9 proteins together, and the AcrIIc3 traps the HNH domain away from the cleavage site, thus preventing DNA cleavage by Cas9. These structures provide insights into Cas9 domain rearrangements, guide-target engagement, cleavage mechanism, and anti-CRISPR inhibition, facilitating the optimization of these genome-editing platforms.
The staffs of the BL-17U1 and BL-19U1 beamlines at the Shanghai Synchrotron Radiation Facility and the BL41XU beamline at SPring-8 have provided technical supports to this work.
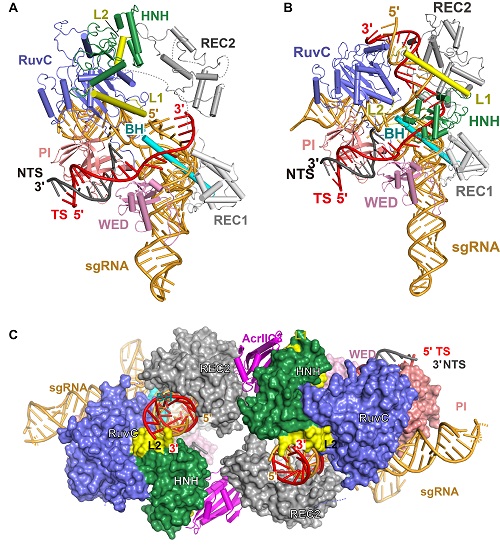
Figure: Structures of seed-matched (A), a fully paired catalytically poised (B), DNA-bound NmeCas9 complexes, and the NmeCas9-sgRNA-DNA-AcrIIC3 complex (C). (Image by Dr. WANG Yanli's group)
