
Circular RNA Formation and Alternative Selection Expand Understanding of Gene Expression Regulation
Sep 23, 2014 Email"> PrintText Size
Published on September 18, 2014 in Cell, the research teams led by Dr. YANG Li from the CAS-MPG Partner Institute for Computational Biology (PICB) and CHEN Lingling from the Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Science (SIBS) of Chinese Academy of Sciences (CAS) demonstrate the mechanism of circular RNA biogenesis and present the generality of alternative circularization for the complexity of mammalian posttranscriptional regulation.
Exon circularization has been identified from many loci in mammals, but the detailed mechanism of its biogenesis has remained elusive. Dr. YANG, Dr. CHEN and their colleagues take advantage of non-polyadenylated and RNase R treated RNA-seq from H9 human embryonic stem cells (hESCs) with a newly developed pipeline to predict back spliced junctions and systematically characterize circular RNAs. Importantly, they have recapitulated circular RNA formation from a unique expression vector, and offer multiple lines of evidence to support the conclusion that circular RNA formation is dependent on flanking complementary sequences, including either repetitive or non-repetitive elements. Strikingly, such sequences exhibit rapid evolutionary changes among mammals, showing that exon circularization is evolutionarily dynamic. Furthermore, they show that the exon circularization efficiency is regulated by the competition of RNA pairing by complementary sequences within individual introns or across flanking introns. Alternative formation of inverted repeated Alu pairs (IRAlus) and the competition between them lead to alternative circularization, resulting in multiple circular RNA transcripts produced from a single gene.
The identification of alternative circularization further expands the understanding of gene expression regulation. Through alternative splicing, multiple functional mRNAs (and proteins) could be produced from a single gene. These multiple functional mRNAs are generally thought to exist only as linearized molecules. This work shows that alternative circularization coupled with alternative splicing can produce a variety of additional circular RNAs from one gene. Collectively, exon circularization mediated by complementary sequences in human introns and the potential to generate alternative circularization products extend the complexity of mammalian posttranscriptional regulation.
This study entitled "Complementary Sequence-Mediated Exon Circularization" was published on Cell as a full article.
This work was supported by grants from Chinese Academy of Sciences, the Ministry of Science and Technology of China, the National Natural Science Foundation of China, and Shanghai Institutes for Biological Sciences, CAS.
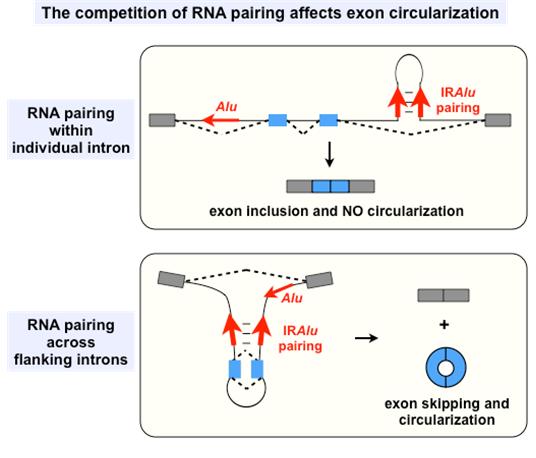
The competition models of RNA pairing by complementary sequence-mediated exon circularization (Image by YANG Li’s research group)
Published on September 18, 2014 in Cell, the research teams led by Dr. YANG Li from the CAS-MPG Partner Institute for Computational Biology (PICB) and CHEN Lingling from the Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Science (SIBS) of Chinese Academy of Sciences (CAS) demonstrate the mechanism of circular RNA biogenesis and present the generality of alternative circularization for the complexity of mammalian posttranscriptional regulation.
Exon circularization has been identified from many loci in mammals, but the detailed mechanism of its biogenesis has remained elusive. Dr. YANG, Dr. CHEN and their colleagues take advantage of non-polyadenylated and RNase R treated RNA-seq from H9 human embryonic stem cells (hESCs) with a newly developed pipeline to predict back spliced junctions and systematically characterize circular RNAs. Importantly, they have recapitulated circular RNA formation from a unique expression vector, and offer multiple lines of evidence to support the conclusion that circular RNA formation is dependent on flanking complementary sequences, including either repetitive or non-repetitive elements. Strikingly, such sequences exhibit rapid evolutionary changes among mammals, showing that exon circularization is evolutionarily dynamic. Furthermore, they show that the exon circularization efficiency is regulated by the competition of RNA pairing by complementary sequences within individual introns or across flanking introns. Alternative formation of inverted repeated Alu pairs (IRAlus) and the competition between them lead to alternative circularization, resulting in multiple circular RNA transcripts produced from a single gene.
The identification of alternative circularization further expands the understanding of gene expression regulation. Through alternative splicing, multiple functional mRNAs (and proteins) could be produced from a single gene. These multiple functional mRNAs are generally thought to exist only as linearized molecules. This work shows that alternative circularization coupled with alternative splicing can produce a variety of additional circular RNAs from one gene. Collectively, exon circularization mediated by complementary sequences in human introns and the potential to generate alternative circularization products extend the complexity of mammalian posttranscriptional regulation.
This study entitled "Complementary Sequence-Mediated Exon Circularization" was published on Cell as a full article.
This work was supported by grants from Chinese Academy of Sciences, the Ministry of Science and Technology of China, the National Natural Science Foundation of China, and Shanghai Institutes for Biological Sciences, CAS.

The competition models of RNA pairing by complementary sequence-mediated exon circularization (Image by YANG Li’s research group)
CAS Institutes
There are 124 Institutions directly under the CAS by the end of 2012, with 104 research institutes, five universities & supporting organizations, 12 management organizations that consist of the headquarters and branches, and three other units. Moreover, there are 25 legal entities affiliated and 22 CAS invested holding enterprisesThere are 124 I...>> more
Contact Us

Chinese Academy of Sciences
Add: 52 Sanlihe Rd., Xicheng District, Beijing, China
Postcode: 100864
Tel: 86-10-68597592 (day) 86-10-68597289 (night)
Fax: 86-10-68511095 (day) 86-10-68512458 (night)
E-mail: cas_en@cas.cn

