
SIMM Develops NUMD for Exploring Transition Pathway and Free-Energy Profile of Large-Scale Protein Conformational Change
Jul 28, 2014 Email"> PrintText Size
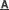
The current view of protein structure now includes a realization that proteins continually undergo complex and hierarchical conformational changes while carrying out their biological functions, e.g., in signal transduction, immune response, protein folding, and enzymatic activity. Large-scale conformational changes of proteins are usually associated with the binding of ligands. Because the conformational changes are often related to the biological functions of proteins, understanding the molecular mechanisms of these motions and the effects of ligand binding becomes very necessary.
The protein conformational transition coupled with the biological function has attracted much attention in the last several decades. Various methods in both experiments and theoretical simulations have been utilized to understand the molecular mechanism of protein conformational transition. However, the difficulty in capturing the transition pathway makes the mechanism unclear yet.
To make a systemic study of the molecular mechanism of protein conformational change and the effects from ligand binding, a research group led by Prof. ZHU Weiliang in Shanghai Institute of Materia Medica (SIMM) combined a normal-mode analysis (NMA) method in internal coordinates (iMOD) and umbrella sampling MD simulation to investigate three typical protein systems: the phosphotransferase enzyme adenylate kinase (AdK) from Escherichia coli, the Ca2+-binding protein calmodulin (CaM), and the p38a mitogen-activated protein kinase (MAPK).
These observations indicate the mechanism diversity of protein conformational change, suggesting that the detailed mechanism of ligand binding and the associated conformational transition is not uniform for all kinds of proteins but may be correlated to their structural flexibility and respective biological functions.
This research work was performed by Dr. WANG Jin’an. The study has been published in the Journal of Physical Chemistry B.

The free energy landscape along the protein conformational change when binding or unbinding ligands (Image By SIMM)
The current view of protein structure now includes a realization that proteins continually undergo complex and hierarchical conformational changes while carrying out their biological functions, e.g., in signal transduction, immune response, protein folding, and enzymatic activity. Large-scale conformational changes of proteins are usually associated with the binding of ligands. Because the conformational changes are often related to the biological functions of proteins, understanding the molecular mechanisms of these motions and the effects of ligand binding becomes very necessary.
The protein conformational transition coupled with the biological function has attracted much attention in the last several decades. Various methods in both experiments and theoretical simulations have been utilized to understand the molecular mechanism of protein conformational transition. However, the difficulty in capturing the transition pathway makes the mechanism unclear yet.
To make a systemic study of the molecular mechanism of protein conformational change and the effects from ligand binding, a research group led by Prof. ZHU Weiliang in Shanghai Institute of Materia Medica (SIMM) combined a normal-mode analysis (NMA) method in internal coordinates (iMOD) and umbrella sampling MD simulation to investigate three typical protein systems: the phosphotransferase enzyme adenylate kinase (AdK) from Escherichia coli, the Ca2+-binding protein calmodulin (CaM), and the p38a mitogen-activated protein kinase (MAPK).
These observations indicate the mechanism diversity of protein conformational change, suggesting that the detailed mechanism of ligand binding and the associated conformational transition is not uniform for all kinds of proteins but may be correlated to their structural flexibility and respective biological functions.
This research work was performed by Dr. WANG Jin’an. The study has been published in the Journal of Physical Chemistry B.

The free energy landscape along the protein conformational change when binding or unbinding ligands (Image By SIMM)
CAS Institutes
There are 124 Institutions directly under the CAS by the end of 2012, with 104 research institutes, five universities & supporting organizations, 12 management organizations that consist of the headquarters and branches, and three other units. Moreover, there are 25 legal entities affiliated and 22 CAS invested holding enterprisesThere are 124 I...>> more
Contact Us

Chinese Academy of Sciences
Add: 52 Sanlihe Rd., Xicheng District, Beijing, China
Postcode: 100864
Tel: 86-10-68597592 (day) 86-10-68597289 (night)
Fax: 86-10-68511095 (day) 86-10-68512458 (night)
E-mail: cas_en@cas.cn

