

A research team led by Dr. LI Guohong and Dr. ZHU Mingzhao from the Institute of Biophysics of the Chinese Academy of Sciences, has demonstrated that the histone variant H2A.Z facilitates the licensing and activation of early DNA replication origins.
Results of the research was published in Nature on Dec. 25th, 2019.
DNA replication is a tightly regulated process that ensures the precise duplication of the genome during cell proliferation. Replication origins determine where replication starts on the genome and regulate the whole genome replication program. The human genome contains tens of thousands of origins; however, only about 10% of them are used in each cell cycle. So how are origins selected?
In eukaryotes, DNA wraps around histone octamers to form chromatin in the nucleus. The licensing and activation of replication origins are regulated by both the DNA sequence and chromatin features. However, chromatin-based regulatory mechanisms remain largely uncharacterized.
In this study, the scientists first found that knocking down H2AFZ genes in HeLa cells results in cell growth defects. Through mass spectrometry, many subunits of prereplication complex were enriched on H2A.Z mono-nucleosomes, indicating that H2A.Z may be involved in the licensing of DNA replication origins.
To find the mechanism of interaction between H2A.Z and the prereplication complex, the scientists then performed in vitro biochemical analysis and found that H2A.Z-containing nucleosomes bind directly to the histone lysine methyltransferase enzyme SUV420H1. This process promotes H4K20me2 deposition, which further recruits origins recognition complex 1 (ORC1) to help accomplish the licensing of DNA replication origins.
In addition, through genome-wide studies in HeLa cells, the scientists confirmed the role of H2A.Z in DNA replication. The results showed that signals from H4K20me2, ORC1 and nascent DNA strands (indicating active DNA replication origins) co-localize with H2A.Z, and the depletion of H2A.Z results in decreased H4K20me2, ORC1 and nascent-strand signals. H2A.Z-regulated replication origins have a higher firing efficiency and earlier replication timing compared with other origins.
Furthermore, the scientists generated CD4CreH2A.Zf/f mice to study the function of H2A.Z-regulated replication in a more physiological context. Using these mice, they conditionally knocked out (CKO) H2az1/H2az2 in T cells. They then found that in H2A.Z CKO mic the activated T cells have defects in cell proliferation and DNA replication.
This study describes a novel epigenetic regulation mechanism for DNA replication origin selection and offers a new way of understanding DNA replication regulation in eukaryotes. Importantly, this regulatory pathway can potentially serve as a target for cancer treatment and regulation of T cell function during immunotherapy.
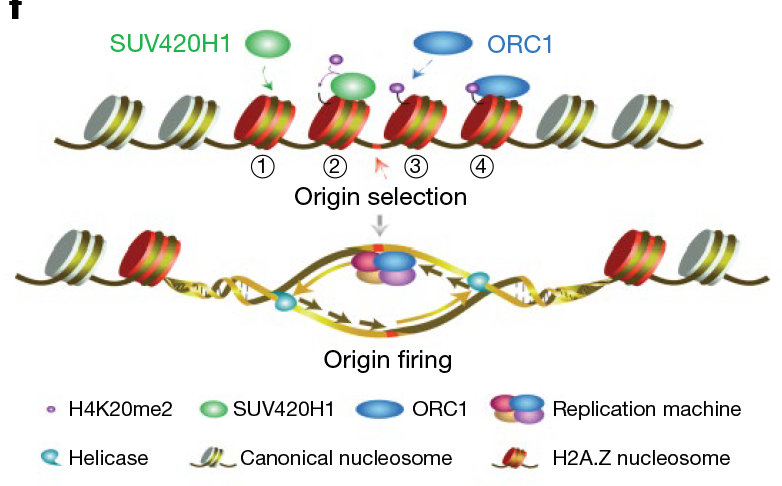
Working model: Origin selection: H2A.Z nucleosomes bind Suv420H1 directly to establish H4K20me2 on chromatin, which then recruits ORC1 to bind to replication origins; Origin firing: H2A.Z-Suv420H1-H4K20me2-ORC1 axis selectively license and activate early replication origins. (Image by Dr. LI Guohong’s lab)