
The epigenomes of mammalian sperm and oocytes, characterized by gamete-specific 5-methylcytosine (5mC) patterns, are reprogrammed during early embryogenesis to establish full developmental potential. A recent study jointly conducted by Profs. XU Guoliang and LI Jinsong's groups from Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Sciences (SIBS) of Chinese Academy of Sciences and Prof. TANG Fuchou's group from Peking University revealed a scenario in early embryonic development of mammals: both paternal and maternal genomes of one-cell mouse embryo (zygote) undergo widespread active and passive DNA demethylation.
Their discovery was different from the earlier belief that only paternal genome undergoes active demethylation while the maternal genome undergoes passive demethylation through DNA replication. This discovery fueled new insights on the role of active and passive DNA demethylation in cell reprogramming and early embryonic development of mammals.
The study also confirmed their earlier discovery that this process of demethylation is mediated by Tet3, a member from the enzyme family of Tet dioxygenases, and is independent of another enzyme, the thymine DNA glycosylase (TDG), which was revealed earlier as a player in the downstream demethylation.
With the aid of high-accuracy analytic techniques for epigenetic modification, including an improved reduced representation bisulfite sequencing approach developed by the researchers that enabled them to determine the methylomes at base resolution on a genome-wide scale, they investigated Tet3- and TDG-knockout mice and directly looked at the pronuclear DNA before the first mitosis in the zygote to test the role of active and passive demethylation. They demonstrated that aside from passive demethylation via DNA replication, both paternal and maternal genomes undergo global active demethylation in the zygote before the first mitotic division.
This demethylation, as reported by the team, is dependent on Tet3. At the loci where active demethylation occurred, the 5mCs were replaced by unmodified cytosines, leaving very little 5fC/5caC, residual products from the Tet-mediated oxidation. The demethylation process is independent of TDG, suggesting existence of other player(s) in the downstream demethylation responsible for eliminating the residuals of 5mC. "This indicates that there may be another demethylation pathway to be identified, in which Tet3, but not TDG is involved." commented Dr. GU Tianpeng, co-author of the paper.
The work entitled “Active and Passive Demethylation of Male and Female Pronuclear DNA in the Mammalian Zygote” has been published in Cell Stem Cell on Oct 2, 2014. The research funding came from the Ministry of Sciences and Technology of China, the National Natural Science Foundation of China, the “Strategic Priority Research Program” of the Chinese Academy of Sciences and the National Science and Technology Major Project of China.
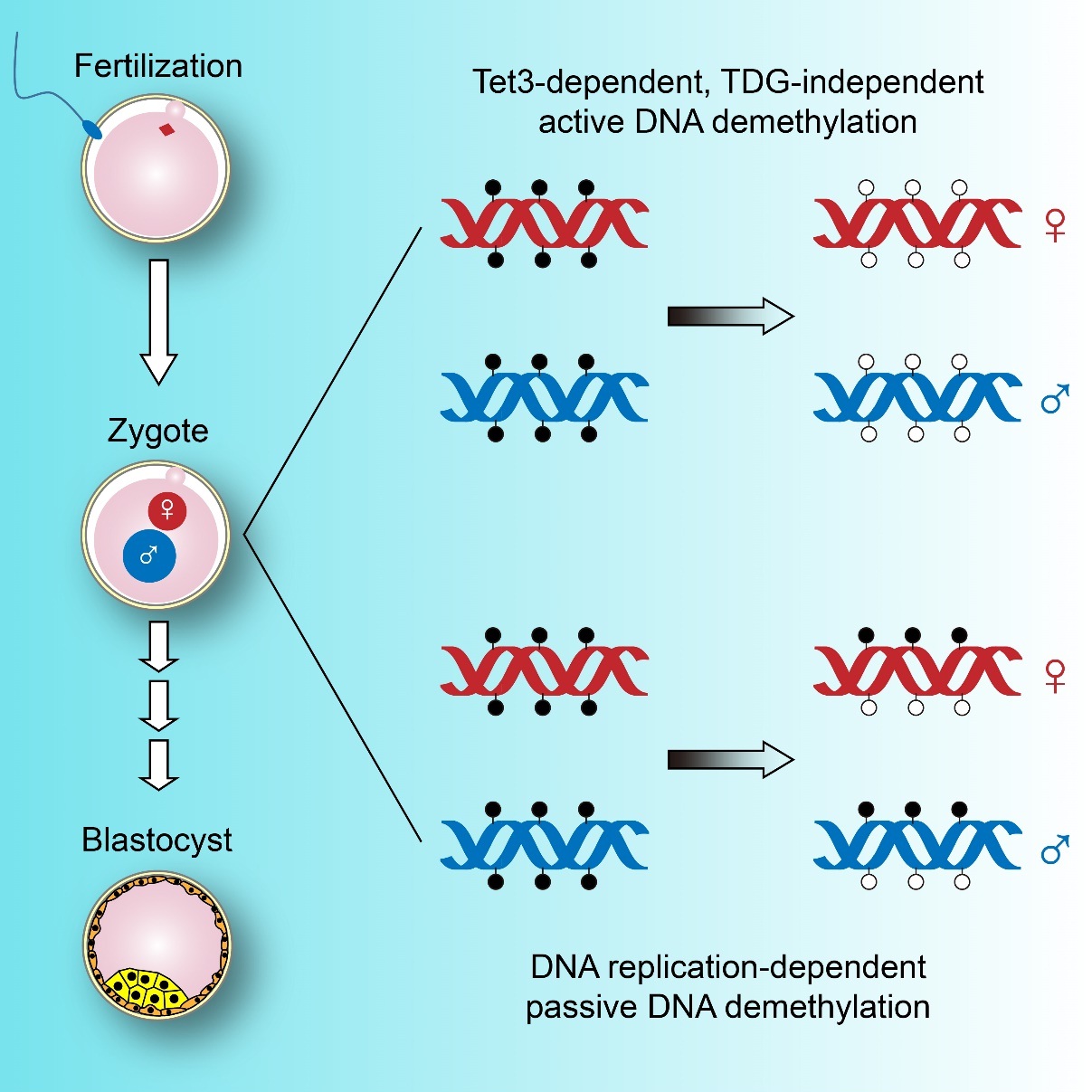
In one-cell mouse embryos, the maternal and paternal genomes both undergo Tet3-dependent active demethylation and replication-mediated passive demethylation. The active demethylation pathway does not require TDG. (Image by XU Guoliang`s group)

86-10-68597521 (day)
86-10-68597289 (night)

86-10-68511095 (day)
86-10-68512458 (night)

cas_en@cas.cn

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

