
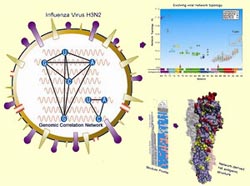
Networks of genomic co-occurrence capture characteristics of human influenza A (H3N2) evolution (Du et al. Genome Research, DOI: 10.1101/gr.6969007)
Understanding the evolution of influenza A virus, which poses a global challenge to public health, is of special significance for its control and prevention. Although the genome structure of the virus is seemingly simple, their evolutionary patterns and molecular mechanisms are difficult to reveal.
The recent availability of full genomic sequence data for a large number of human influenza A (H3N2) virus isolates over many years have provided an opportunity to analyze its evolution by considering all gene segments simultaneously. However, such analysis requires new computational models that can capture the complex evolutionary features over the entire genome.
As being reported online by Genome Research on 21 November, a research group headed by Prof. JIANG Taijiao with the CAS Institute of Biophysics has set up a new computational model to depict many epidemiological characteristics of the A (H3N2) virus.
By analyzing the co-occurrence of the nucleotides in the entire genome of the virus, Prof. Jiang and colleagues have developed a network model to describe its evolutionary patterns and dynamics. The network model can effectively identify the antigenic features of the virus at the whole-genome level and accurately distinguish the complex patterns in the viral evolution between different gene segments. Their work further shows that the co-occurring nucleotide modules apparently underpin the dynamics of the H3N2 viral evolution. Moreover, the network model allowed them to identify key amino acid substitutions that dictate the antigenic evolution of human H3N2 virus. The study demonstrates that the network of nucleotide co-occurrences presents a promising method for tracking down the route of the viral evolution.
"The idea that a network can model its evolution is a streamlined approach to further understand and predict the patterns of global epidemics and will be a great step forward in virus biology," according to a reviewer of the paper. This work is also highly appreciated by another reviewer, "Few papers on novel methods for the analysis of large collections of influenza genomes have been published to date, and this paper represents a significant advance."

86-10-68597521 (day)
86-10-68597289 (night)

86-10-68511095 (day)
86-10-68512458 (night)

cas_en@cas.cn

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

